7 Interaction Analysis Nichenet
Florian Wuenenmann
4/12/2022
Last updated: 2022-04-19
Checks: 7 0
Knit directory: Abatacept_scrnaseq_mi/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220318) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 4d0c7b6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: code/utils.R
Untracked: code/utils_nichenet.R
Untracked: data/Table_1_Common Transcriptomic Effects of Abatacept and Other DMARDs on Rheumatoid Arthritis Synovial Tissue.xlsx
Untracked: omnipathr-log/
Unstaged changes:
Deleted: analysis/7_Conclusions.Rmd
Deleted: analysis/7_summary.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/7_interaction_analysis_nichenet.Rmd) and HTML (docs/7_interaction_analysis_nichenet.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 4d0c7b6 | Florian Wuennemann | 2022-04-19 | Main Update to code and adding NichenetR |
| html | 068439f | Florian Wuennemann | 2022-04-19 | Build site. |
| Rmd | dcc5db0 | Florian Wuennemann | 2022-04-19 | Main Update to code and adding NichenetR |
| html | 48c189a | Florian Wuennemann | 2022-04-19 | Build site. |
| Rmd | 593b491 | Florian Wuennemann | 2022-04-19 | Main Update to code and adding NichenetR |
| html | fc663eb | Florian Wuennemann | 2022-04-19 | Build site. |
| Rmd | 046d56c | Florian Wuennemann | 2022-04-19 | Main Update to code and adding NichenetR |
| html | 8d9ac5d | Florian Wuennemann | 2022-04-19 | Build site. |
| html | a71b19c | Florian Wuennemann | 2022-04-19 | Build site. |
| Rmd | 86ff3e7 | Florian Wuennemann | 2022-04-19 | Main Update to code and adding NichenetR |
Aim
In this markdown, we will perform cell-interaction analysis to try to identify critical ligands and receptors that are involved in the transcriptional response to Abatacept treatment. For this, we will use LIANA and Nichenet.
The questions we want to answer with this approach is: - Which ligands expressed by immune cells are most likely to induce the upregulation of inflammatory response genes in activated fibroblasts?
- Which ligands expressed/not expressed anymore by antigen presenting cells treated with Abatacept induce specific gene programs in T-cells or Macrophages.
More specific follow-up analysis can be performed according to the tutorials by the Nichenet authors:
https://github.com/saeyslab/nichenetr
In this short analysis, I am simply using Nichenet directly, but more integrated analysis with Omnipath and LIANA can be performed.
Load Seurat object
Let#s load our processed Seurat object first.
# Annotated seurat object
seurat_object <- readRDS(here("../data/2_annotated.seurat_object.rds"))Manually check expression of candidate ligands and receptors for Abatacept
Let’s check the expression of the Ctl4a gene, which should be expressed pretty specifically in T-cells. We also check the expression of the canonical Ctl4a receptors Cd80/Cd86
## Let's check expression of CD80, CD86 and CTL4A in all cell types
DotPlot(seurat_object, features = c("Cd80","Cd86","Ctla4"), split.by = "group") + RotatedAxis()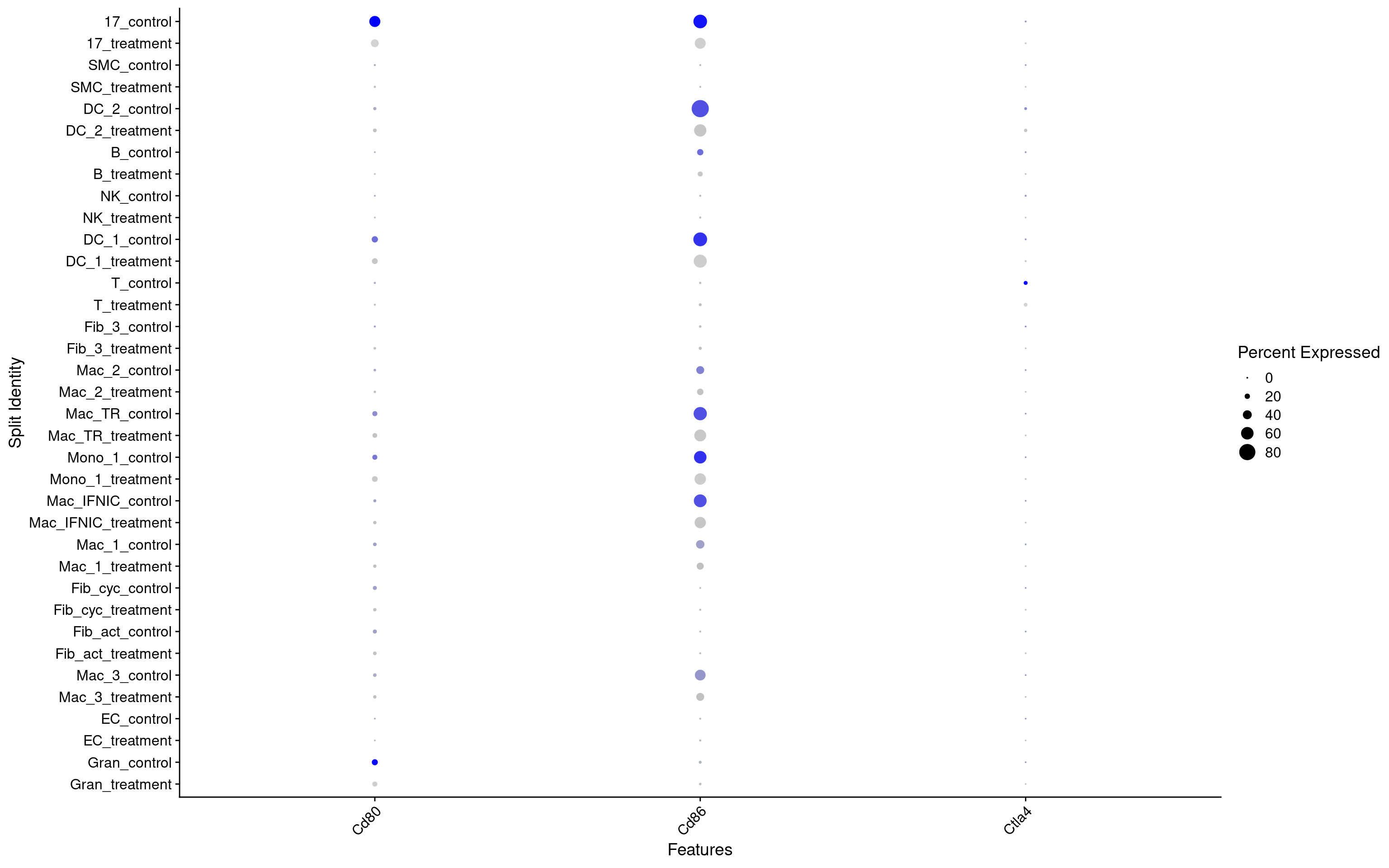
The data confirms what we were expecting beforehand, that Ctl4a is specifically expressed by T-cells. The receptors Cd80/Cd86 seem to be expressed in many cell types, with Cd86 being expressed by more cells than Cd80, which seems specific to cluster 17, DC_1 and Granulocytes.
Nichenet
Now we will perform a Nichenet analysis, in which we will try to identify ligand-receptor interactions, that can explain gene expression differences in receiver cells between conditions.
Download reference data
First, we will run a specific analysis, to see which ligand-receptor interactions are most likely to explain gene-expression differences in T-cells, since Abatacept is supposed to inhibit T-cell activation.
As a first step, we download the Nichenet reference data.
# Only download this file once, then store it locally
nichenet_human <- here("../references/ligand_target_matrix.rds")
# if(file.exists(nichenet_human)){
ligand_target_matrix <- readRDS(nichenet_human)
# }else{
# ligand_target_matrix = readRDS(url("https://zenodo.org/record/3260758/files/ligand_target_matrix.rds"))
# saveRDS(ligand_target_matrix,nichenet_human)
# }## Load other references for NichenetR
lr_network = readRDS(here("../references/lr_network.rds"))
weighted_networks = readRDS(here("../references/weighted_networks.rds"))Load DE gene and pathway list
We will load both DE gene tables and pathway analysis results. Since the DE gene power is very limited, we may want to focus on the pathway resultsa bit more.
de_genes <- fread(here("../results/3_DE_genes.merged_wilcox_pseudobulk.tsv"))
de_paths <- fread(here("../results/pathway_enrichment.GSEA_Hallmark.all_res.tsv"))
gene.sets <- getGeneSets(species = "Mus musculus", library = c("H"))Activated Fibroblast receiver, all other cells sender
nichenet_fibact_obj <- here("../results/nichenet_fibact.object.rds")
if(!file.exists(nichenet_fibact_obj)){
nichenet_output_fib_act = nichenet_seuratobj_aggregate(
seurat_obj = seurat_object,
receiver = c("Fib_act"),
condition_colname = "group",
condition_oi = "treatment", condition_reference = "control",
sender = "all",
ligand_target_matrix = ligand_target_matrix, lr_network = lr_network,
weighted_networks = weighted_networks,
organism = "mouse")
saveRDS(nichenet_output_fib_act,
file = nichenet_fibact_obj)
}else{
nichenet_output_fib_act <- readRDS( nichenet_fibact_obj)
}
# Plot results
nichenet_output_fib_act$ligand_activity_target_heatmap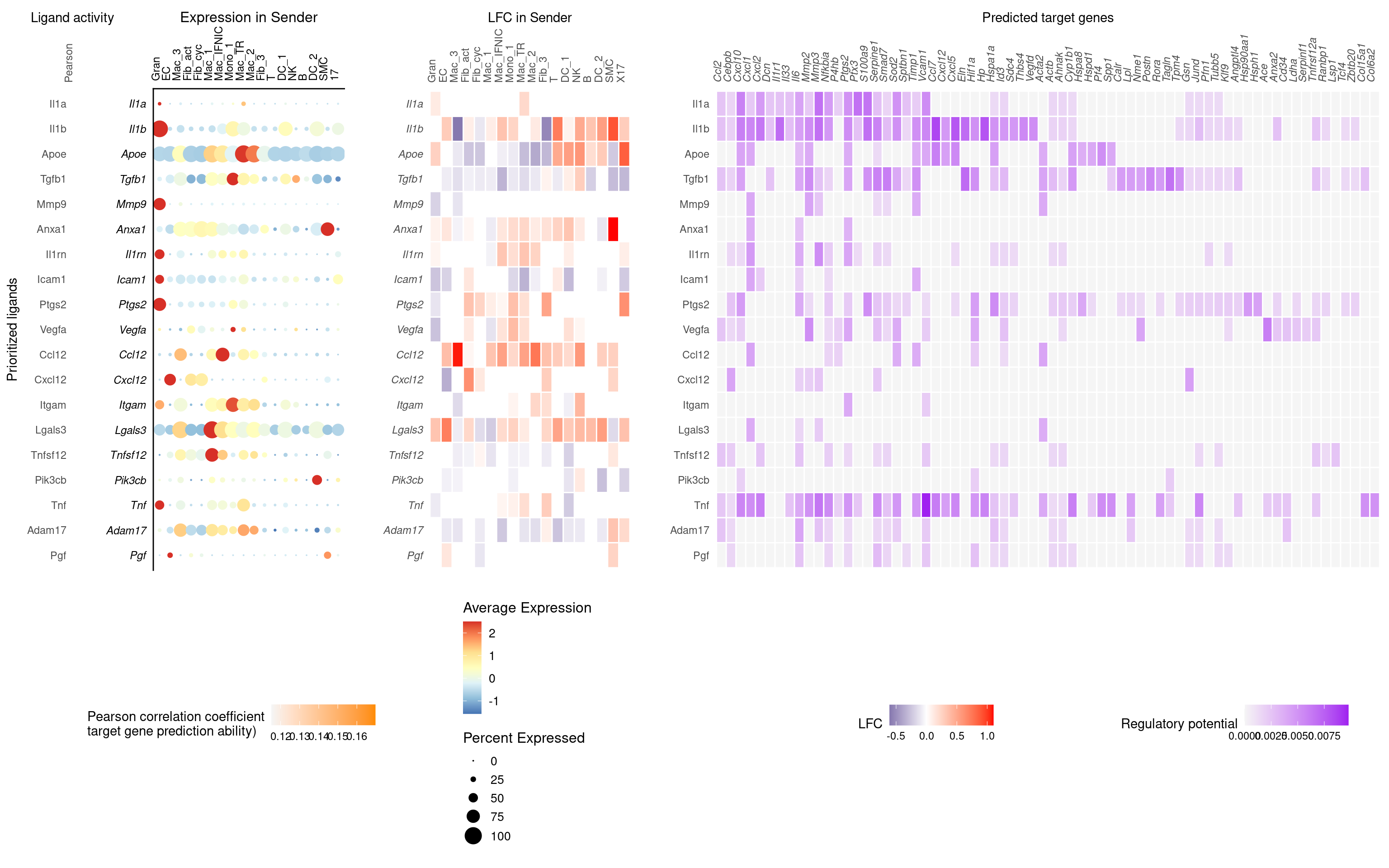
DotPlot(seurat_object, features = c("Il1a","Il1b","Apoe","Mmp9"), split.by = "group") + RotatedAxis()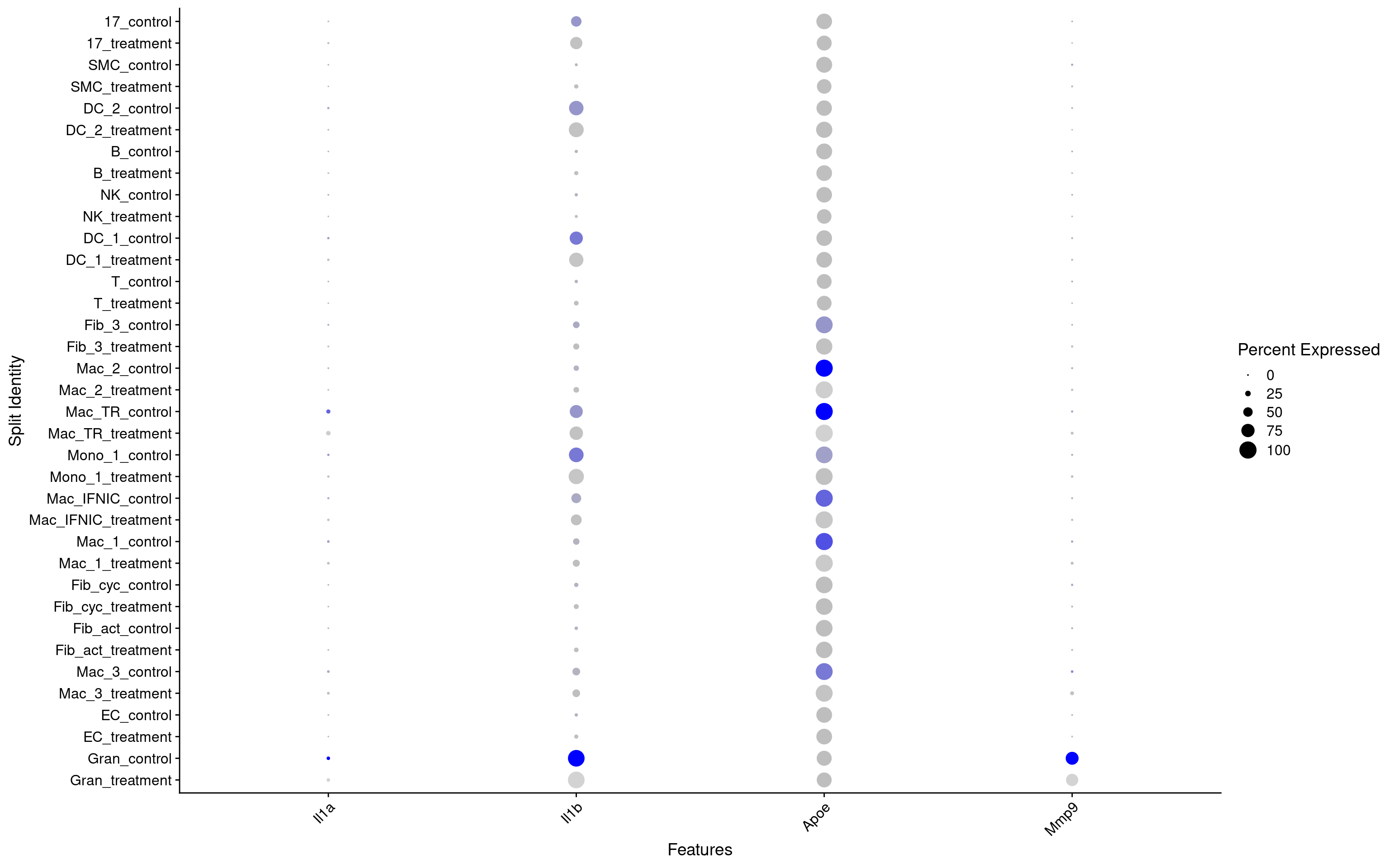
T-cells receiver, all other cells sender
nichenet_tcell_obj <- here("../results/nichenet_tcell.object.rds")
if(!file.exists(nichenet_tcell_obj)){
nichenet_output_tcell = nichenet_seuratobj_aggregate(
seurat_obj = seurat_object,
receiver = c("T"),
condition_colname = "group",
condition_oi = "treatment", condition_reference = "control",
sender = "all",
ligand_target_matrix = ligand_target_matrix,
lr_network = lr_network,
weighted_networks = weighted_networks,
organism = "mouse")
saveRDS(nichenet_output_tcell,
file = nichenet_tcell_obj)
}else{
nichenet_output_tcell <- readRDS( nichenet_tcell_obj)
}
nichenet_output_tcell$ligand_activity_target_heatmap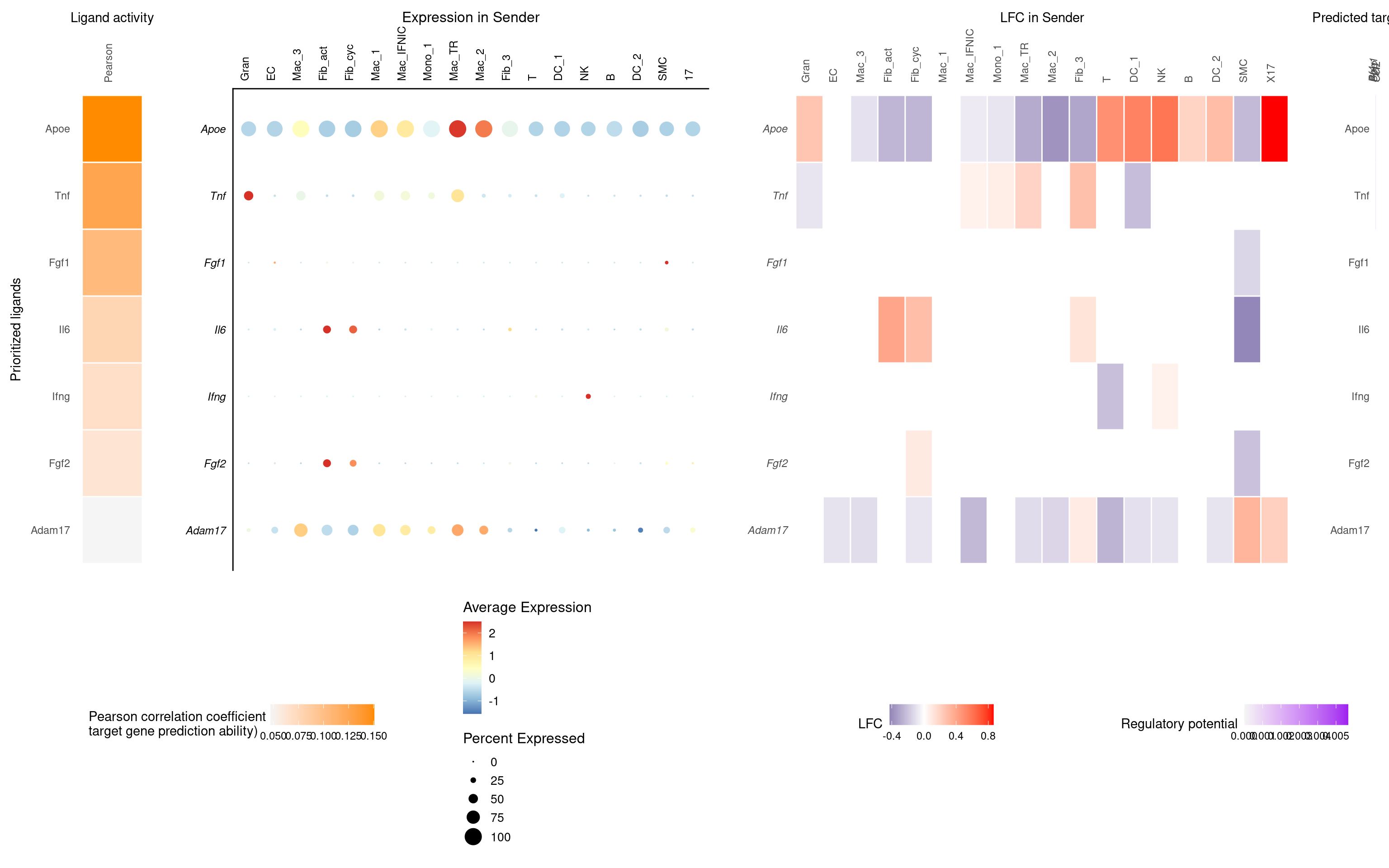
DotPlot(seurat_object, features = c("Apoe","Tnf","Fgf1","Pf4","Spp1"), split.by = "group") + RotatedAxis()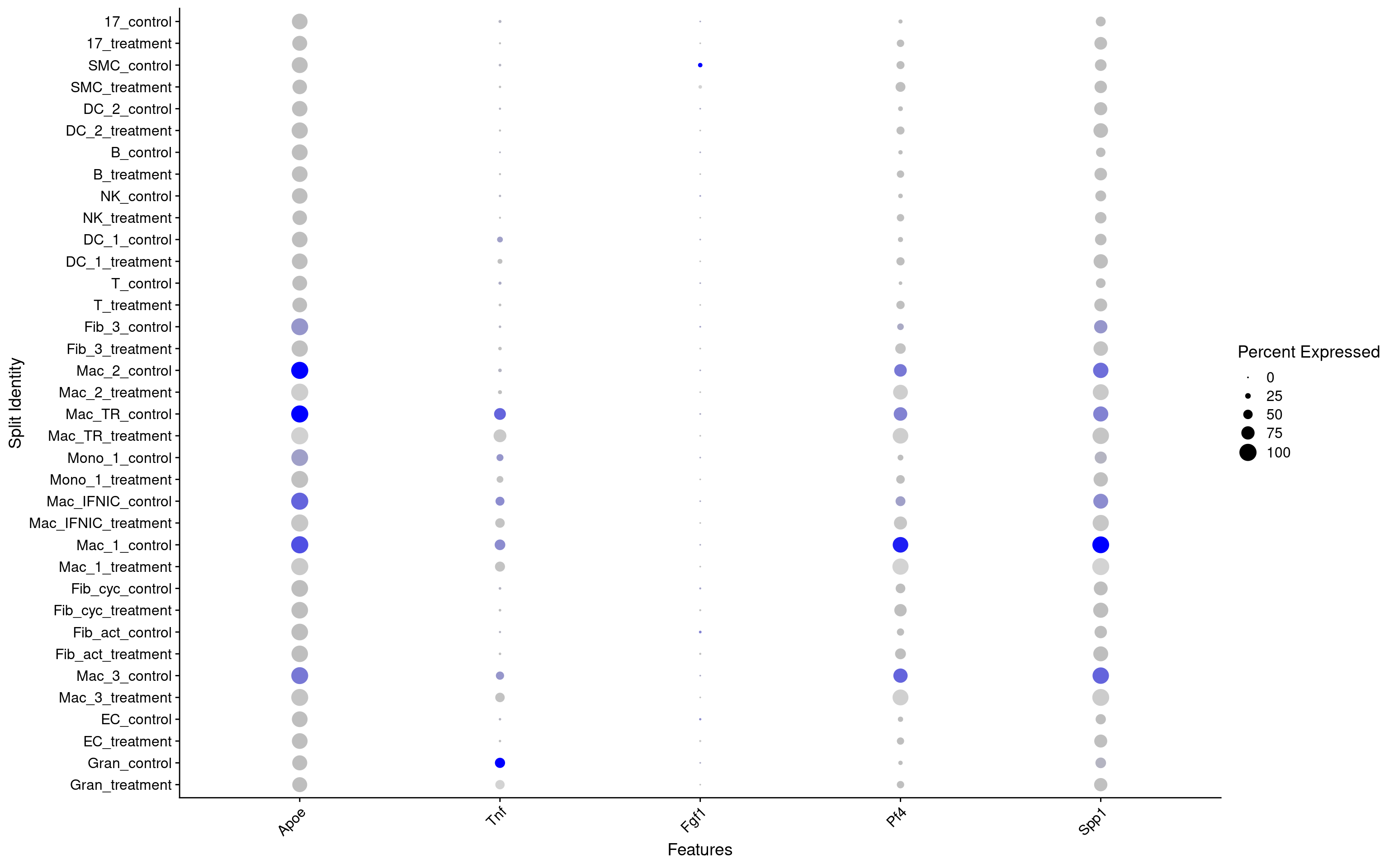
sessionInfo()R version 4.1.3 (2022-03-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=de_DE.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=de_DE.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=de_DE.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=de_DE.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] RColorBrewer_1.1-3 escape_1.4.1 data.table_1.14.2
[4] nichenetr_1.1.0 here_1.0.1 ComplexHeatmap_2.10.0
[7] ggrepel_0.9.1 SeuratObject_4.0.4 Seurat_4.1.0
[10] magrittr_2.0.3 biomaRt_2.51.3 forcats_0.5.1
[13] stringr_1.4.0 dplyr_1.0.8 purrr_0.3.4
[16] readr_2.1.2 tidyr_1.2.0 tibble_3.1.6
[19] ggplot2_3.3.5 tidyverse_1.3.1 liana_0.0.9
[22] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] rsvd_1.0.5 Hmisc_4.6-0
[3] ica_1.0-2 class_7.3-20
[5] ps_1.6.0 foreach_1.5.2
[7] lmtest_0.9-40 rprojroot_2.0.3
[9] crayon_1.5.1 spatstat.core_2.4-2
[11] MASS_7.3-56 rhdf5filters_1.6.0
[13] nlme_3.1-157 backports_1.4.1
[15] reprex_2.0.1 rlang_1.0.2
[17] XVector_0.34.0 caret_6.0-91
[19] ROCR_1.0-11 readxl_1.4.0
[21] irlba_2.3.5 callr_3.7.0
[23] limma_3.50.3 filelock_1.0.2
[25] BiocParallel_1.28.3 rjson_0.2.21
[27] bit64_4.0.5 glue_1.6.2
[29] sctransform_0.3.3 parallel_4.1.3
[31] processx_3.5.3 spatstat.sparse_2.1-0
[33] AnnotationDbi_1.56.2 BiocGenerics_0.40.0
[35] spatstat.geom_2.4-0 haven_2.4.3
[37] tidyselect_1.1.2 SummarizedExperiment_1.24.0
[39] UCell_1.3.1 fitdistrplus_1.1-8
[41] XML_3.99-0.9 zoo_1.8-9
[43] xtable_1.8-4 evaluate_0.15
[45] scuttle_1.4.0 cli_3.2.0
[47] zlibbioc_1.40.0 rstudioapi_0.13
[49] miniUI_0.1.1.1 whisker_0.4
[51] logger_0.2.2 bslib_0.3.1
[53] rpart_4.1.16 shiny_1.7.1
[55] GSVA_1.42.0 BiocSingular_1.10.0
[57] xfun_0.30 clue_0.3-60
[59] cluster_2.1.3 caTools_1.18.2
[61] KEGGREST_1.34.0 listenv_0.8.0
[63] Biostrings_2.62.0 png_0.1-7
[65] future_1.24.0 ipred_0.9-12
[67] withr_2.5.0 bitops_1.0-7
[69] plyr_1.8.7 cellranger_1.1.0
[71] GSEABase_1.56.0 hardhat_0.2.0
[73] e1071_1.7-9 dqrng_0.3.0
[75] pROC_1.18.0 pillar_1.7.0
[77] GlobalOptions_0.1.2 cachem_1.0.6
[79] fs_1.5.2 flexmix_2.3-17
[81] GetoptLong_1.0.5 DelayedMatrixStats_1.16.0
[83] vctrs_0.4.1 ellipsis_0.3.2
[85] generics_0.1.2 lava_1.6.10
[87] tools_4.1.3 foreign_0.8-82
[89] munsell_0.5.0 proxy_0.4-26
[91] DelayedArray_0.20.0 fastmap_1.1.0
[93] compiler_4.1.3 abind_1.4-5
[95] httpuv_1.6.5 plotly_4.10.0
[97] GenomeInfoDbData_1.2.7 prodlim_2019.11.13
[99] gridExtra_2.3 edgeR_3.36.0
[101] lattice_0.20-45 deldir_1.0-6
[103] visNetwork_2.1.0 utf8_1.2.2
[105] later_1.3.0 BiocFileCache_2.2.1
[107] recipes_0.2.0 SeuratWrappers_0.3.0
[109] jsonlite_1.8.0 scales_1.2.0
[111] graph_1.72.0 ScaledMatrix_1.2.0
[113] pbapply_1.5-0 sparseMatrixStats_1.7.0
[115] lazyeval_0.2.2 promises_1.2.0.1
[117] doParallel_1.0.17 R.utils_2.11.0
[119] latticeExtra_0.6-29 goftest_1.2-3
[121] spatstat.utils_2.3-0 reticulate_1.24
[123] checkmate_2.0.0 rmarkdown_2.13
[125] cowplot_1.1.1 statmod_1.4.36
[127] Rtsne_0.15 Biobase_2.54.0
[129] uwot_0.1.11 igraph_1.3.0
[131] HDF5Array_1.22.1 survival_3.2-13
[133] yaml_2.3.5 htmltools_0.5.2
[135] memoise_2.0.1 modeltools_0.2-23
[137] locfit_1.5-9.5 IRanges_2.28.0
[139] viridisLite_0.4.0 digest_0.6.29
[141] assertthat_0.2.1 mime_0.12
[143] rappdirs_0.3.3 RSQLite_2.2.12
[145] future.apply_1.8.1 remotes_2.4.2
[147] blob_1.2.3 R.oo_1.24.0
[149] S4Vectors_0.32.4 DiagrammeR_1.0.9
[151] labeling_0.4.2 splines_4.1.3
[153] Formula_1.2-4 Rhdf5lib_1.16.0
[155] RCurl_1.98-1.6 broom_0.8.0
[157] hms_1.1.1 modelr_0.1.8
[159] rhdf5_2.38.1 colorspace_2.0-3
[161] base64enc_0.1-3 BiocManager_1.30.16
[163] GenomicRanges_1.46.1 shape_1.4.6
[165] nnet_7.3-17 sass_0.4.1
[167] Rcpp_1.0.8.3 RANN_2.6.1
[169] circlize_0.4.14 fansi_1.0.3
[171] tzdb_0.3.0 parallelly_1.31.0
[173] ModelMetrics_1.2.2.2 R6_2.5.1
[175] ggridges_0.5.3 lifecycle_1.0.1
[177] bluster_1.4.0 curl_4.3.2
[179] leiden_0.3.9 jquerylib_0.1.4
[181] Matrix_1.4-1 RcppAnnoy_0.0.19
[183] iterators_1.0.14 gower_1.0.0
[185] htmlwidgets_1.5.4 beachmat_2.10.0
[187] polyclip_1.10-0 rvest_1.0.2
[189] mgcv_1.8-40 globals_0.14.0
[191] htmlTable_2.4.0 patchwork_1.1.1
[193] spatstat.random_2.2-0 codetools_0.2-18
[195] matrixStats_0.61.0 lubridate_1.8.0
[197] metapod_1.2.0 randomForest_4.7-1
[199] getPass_0.2-2 prettyunits_1.1.1
[201] SingleCellExperiment_1.16.0 dbplyr_2.1.1
[203] R.methodsS3_1.8.1 GenomeInfoDb_1.30.1
[205] gtable_0.3.0 DBI_1.1.2
[207] git2r_0.30.1 stats4_4.1.3
[209] highr_0.9 tensor_1.5
[211] httr_1.4.2 KernSmooth_2.23-20
[213] stringi_1.7.6 progress_1.2.2
[215] farver_2.1.0 reshape2_1.4.4
[217] msigdbr_7.5.1 annotate_1.72.0
[219] fdrtool_1.2.17 timeDate_3043.102
[221] xml2_1.3.3 BiocNeighbors_1.12.0
[223] OmnipathR_3.3.21 scattermore_0.8
[225] scran_1.22.1 bit_4.0.4
[227] jpeg_0.1-9 MatrixGenerics_1.7.0
[229] spatstat.data_2.1-4 babelgene_22.3
[231] pkgconfig_2.0.3 knitr_1.38